Authors: Mark A. Arick II, Corrinne E. Grover, Chuan-Yu Hsu, Zenaida Magbanua, Olga Pechanova, Emma R. Miller, Adam Thrash, Ramey C. Youngblood, Lauren Ezzell, Md. Samsul Alam, John A. H. Benzie, Matthew G. Hamilton, Attila Karsi, Mark L. Lawrence, and Daniel G. Peterson
Abstract
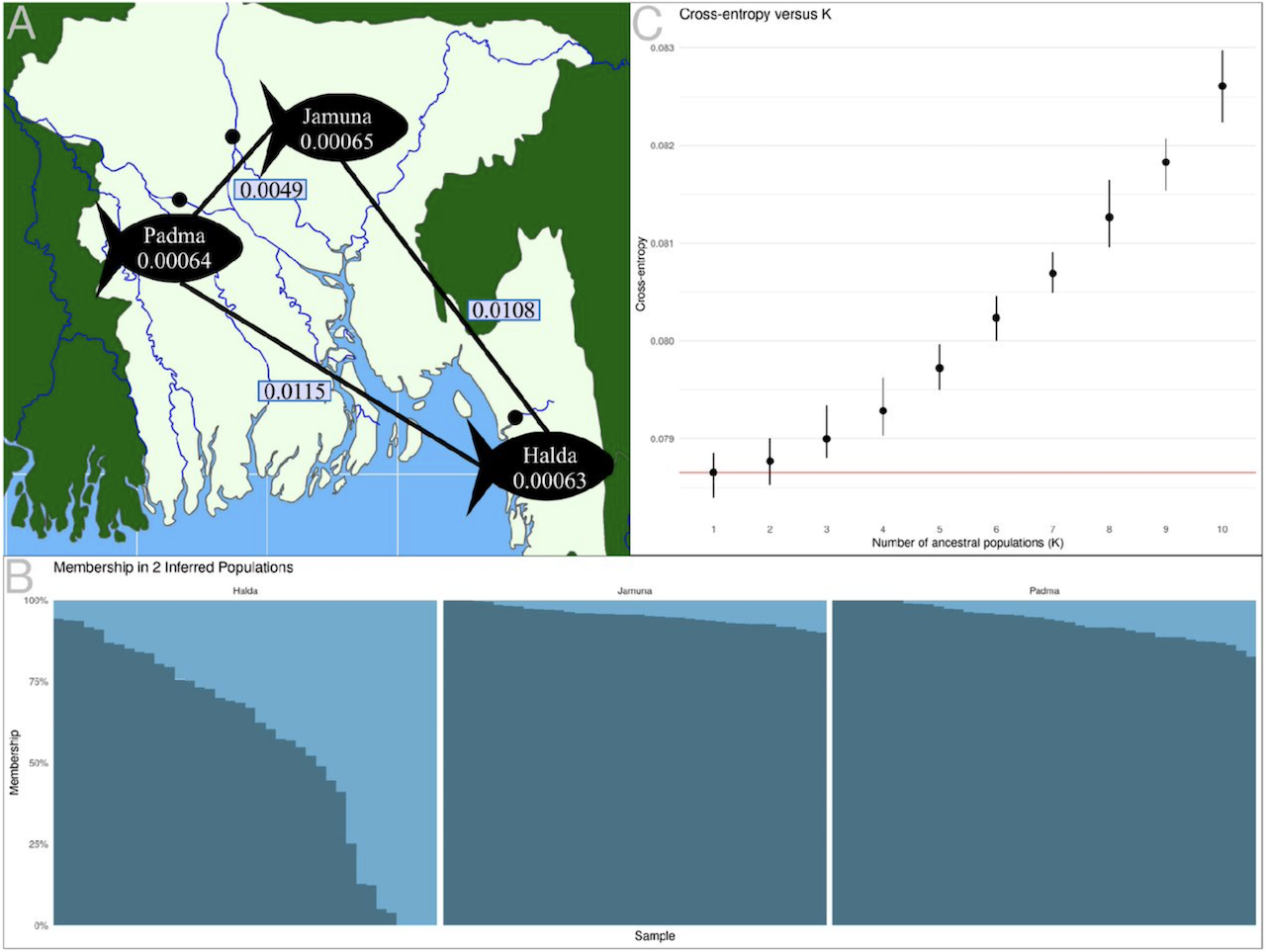
Labeo rohita (rohu) is a carp important to aquaculture in South Asia, with a production volume close to Atlantic salmon. While genetic improvements to rohu are ongoing, the genomic methods commonly used in other aquaculture improvement programs have historically been precluded in rohu, partially due to the lack of a high quality reference genome. Here we present a high-quality de novo genome produced using a combination of next-generation sequencing technologies, resulting in a 946 Mb genome consisting of 25 chromosomes and 2,844 unplaced scaffolds. Notably, while approximately half the size of the existing genome sequence, our genome represents 97.9% of the genome size newly estimated here using flow cytometry. Sequencing from 120 individuals was used in conjunction with this genome to predict the population structure, diversity, and divergence in three major rivers (Jamuna, Padma, and Halda), in addition to infer a likely sex determination mechanism in rohu. These results demonstrate the utility of the new rohu genome in modernizing some aspects of rohu genetic improvement programs.
Read the full publication at https://www.biorxiv.org/content/10.1101/2022.09.08.507226v1.full or at https://doi.org/10.1093/g3journal/jkad009.
Published October 11, 2022

